Graduate Student Researcher, Aug 2015 – Feb 2022 (6 years)
University of Pennsylvania, Philadelphia, PA
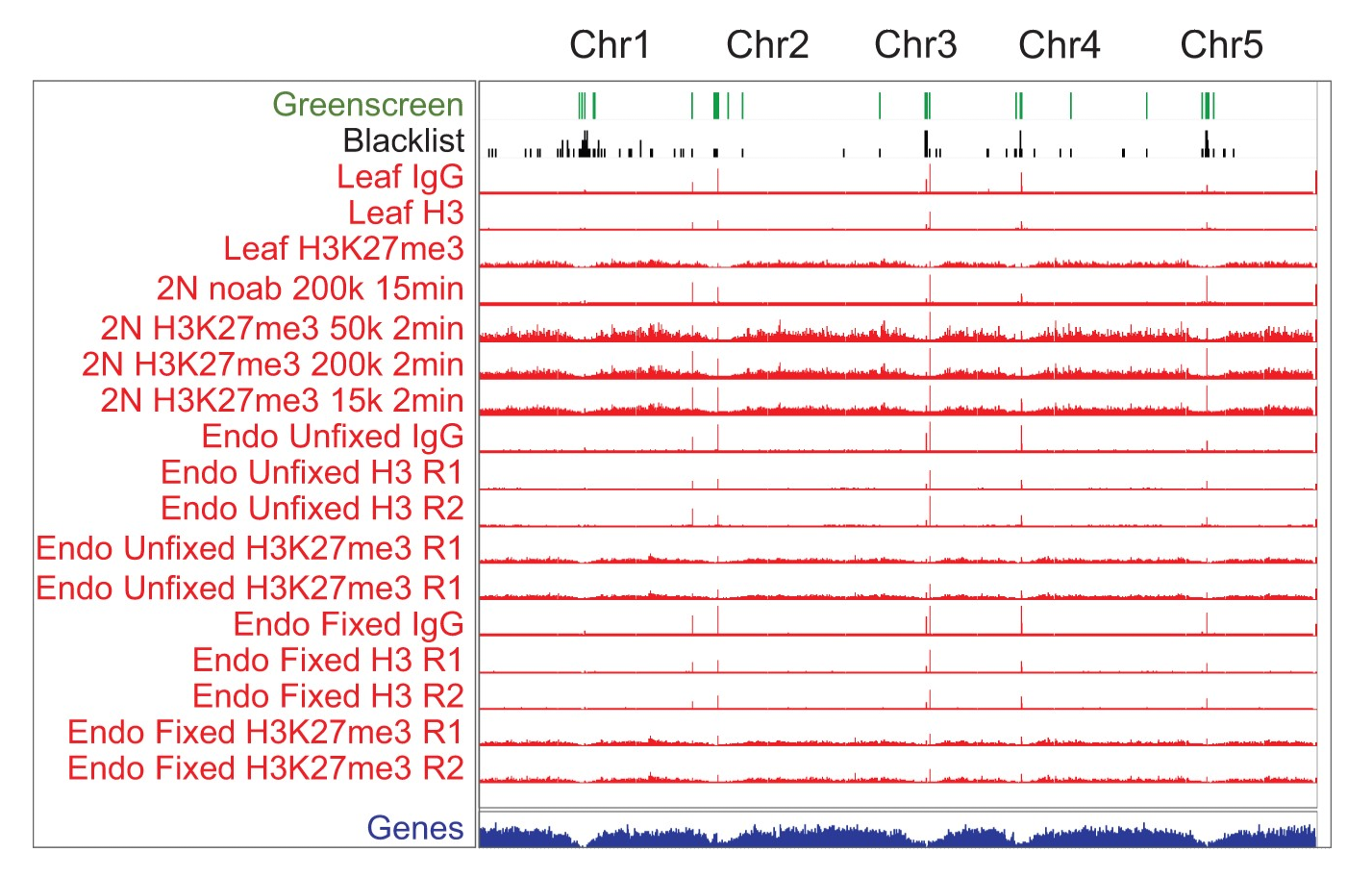
Figure. Cut & Run reads mapped to Arabidopsis genome show that greenscreen regions identify regions of ultra-high artifacts in both ChIP-seq and Cut & Run sequencing experiments (Klasfeld et al 2022).
Summary:
-
Developed Greenscreen method to accurately identify and mask false-positive ChIP-seq peaks with efficacy in Arabidopsis, Drosophila, and Oryza sativa resulting in 1st author publication
-
Designed and orchestrated Next Generation Sequencing (NGS) data analysis (eg. ChIP-Seq, RNA-seq, MNase-seq) to measure the activity and outcomes of mobile and DNA binding proteins functional in cellular reprogramming, resulting in three 2nd author publications
Publications
- Klasfeld, S., & Wagner, D. (2022). Greenscreen decreases Type I Errors and increases true peak detection in genomic datasets including ChIP-seq. bioRxiv.
- Zhu, Y., Klasfeld, S., & Wagner, D. (2021). Molecular regulation of plant developmental transitions and plant architecture via PEPB family proteins–an update on mechanism of action. Journal of Experimental Botany.
- Jin, R., Klasfeld, S., … & Wagner, D., (2021). LEAFY is a pioneer transcription factor and licenses cell reprogramming to floral fate. Nature communications, 12(1), 1-14.
- Zhu, Y., Klasfeld, S., …, & Wagner, D. (2020). TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nature communications, 11(1), 1-12
- Xiao, J., …, Klasfeld, S., …, & Wagner, D., (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nature genetics, 49(10), p.1546.
Presentations
- Klasfeld, S., & Wagner, D. (2019, June 12). Improving resolution of protein binding sites by filtering conserved ultra-high signal in Arabidopsis [Poster]. University of Pennsylvania Genomics and Computational Biology Graduate Group Retreat, The College of Physicians of Philadelphia, Philadelphia, PA.
- Klasfeld, S., & Wagner, D. (2018, November 15). Resolving the mechanism of PRC2 during plant stress [Poster]. University of Pennsylvania Epigenetics Retreat, Independence Seaport Museum, Philadelphia, PA.
- Klasfeld, S., & Wagner, D. (2017, October 13). The characterization and discovery of novel Polycomb and Trithorax Recruitment Elements in plants [Poster]. University of Pennsylvania Epigenetics Retreat, Citizens Bank Park, Philadelphia, PA.
Selected Awards
- Honorable Mention of the 2016 National Science Foundation (NSF) Graduate Research Fellowship
References
- Lab website: https://web.sas.upenn.edu/wagner-lab/